
Phenyx ®
Program Reviewed: Phenyx ®
Version Reviewed: Phenyx ®
Date Reviewed: 11/20/2005
Vendor: GeneBio
Web site: http://www.phenyx-ms.com
Use: Proteomics, Comparative Proteomics, Protein Identification
Cost: Phenyx is free on the web, GeneBio does sell a fully functional standalone local version, similar to the way Matrix Science has a limited on-line version and a fully functional standalone version for sale.
Fully Functional Demo Available for Download: Accessible on the web
Log In: Create A Free Account
Contact Person at GeneBio: Olivier Philippe, olivier.philippe@genebio.com
Please Note: Phenyx ® is a registered trademark of Geneva Bioinformatics (GeneBio) SA., Mascot ® is a registered trademark of Matrix Science Ltd., X!Tandem was created by Craig and Beavis of the Manitoba Proteomics Center
Introduction:
Phenyx is a new sequence search engine that was released in September of 2004 based on the true probabilistic and flexible scoring system OLAV. It uses mass spectrometry data to identify proteins. Phenyx provides an automated second round search function. The automated second round search works like this, the first round search identifies the main proteins in the sample set, then the second round search, searches only those proteins with a larger variable set to catch a broad range of post-translational modifications, non tryptic like cleavages, etc. This function makes comprehensive proteomic searches much faster. The on-line version of Phenyx allows you to search uniprot_sprot, uniprot_tremble, and NCBInr sequence databases. One of the unique features is the ability to search multiple databases in a single search! We have found that the NCBInr database is fairly comprehensive and adequate for most searches. Phenyx accepts data in the following formats: mgf, dta, pkl, btdx, mzdata, and mzxml. You can use the free on-line version of Phenyx as a guest user, however we would recommend that you create a free account so that you can save your results and make modifications to your account and searching strategies. Other unique features of Phenyx include the ability to compare multiple proteomic searches side by side (comparison of Phenyx runs together, as well as the option to import Mascot and SEQUEST results!) and the ability to define new mods and enzymes and save them for later searches. There are additional features in Phenyx that can be very handy, for example the AC list box, which allows one to search one or more protein entries for a targeted investigation. We also like the fact that the mass shift is included next to the modification in the fixed and variable mod lists, this additional identifier makes it easier to pick the mod. Another valuable aspect of the Phenyx web portal is the ability to submit up to four search jobs simultaneously!
Below is a peek at what you will see when you try Phenyx. We have included screen shots of the various menus as we submitted a proteomics file for database searching.
Phenyx Web Version Functional Review
(If you would like to follow along you can download the proteomic data file, (1,679 KB), right click and choose "save as" to save the data file to your computer. If you want to follow along you will also need to go to the Phenyx website, and create an account. It only takes a few minutes to receive a password protected account)
Figure 1.
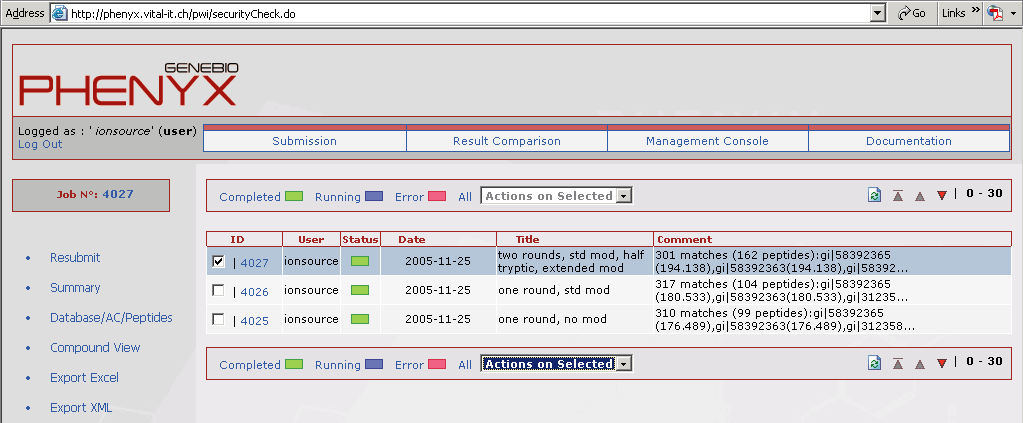
Figure 1. This screen shot shows what the home page of your account will look like. All of the jobs that you submitted will be listed here. Access to the submission page, results comparison page, and management console are accessed through buttons on your account home page.
Figure 2.
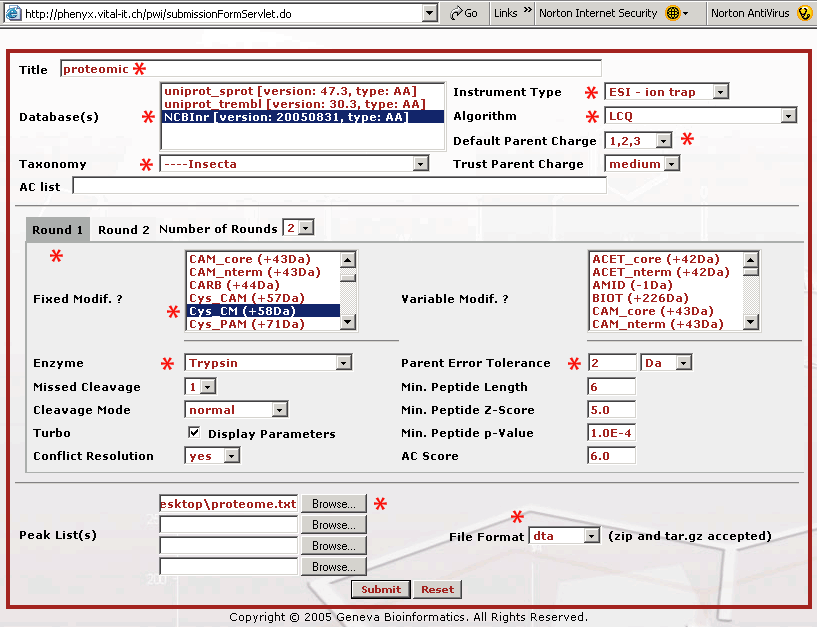
Figure 2. This is the submission page. We have starred all of the fields that we needed to change on the Round 1 search page when we submitted the data file, proteome.dta . As you are filling out the form it is recommended that you name your submission so the search will make sense to you later when you see it in your archive. The database that we will search is the NCBInr protein database, and since this was an insect proteome we selected "insecta" as the taxonomy. The mass spectrometer used was an LCQ XP Plus ion trap mass spectrometer. We made no measurement of the charge state so we picked "Default Parent Charge 1,2,3" and we selected Trust Parent Charge = medium to allow Phenyx taking in account all combinations (that means two charges are considered to be one or two charges). The cysteines in the sample were reduced and S-carboxymethylated with iodoacetic acid, "Cys CM (+58Da)" as part of the sample work up (thus selected as fixed modification). Trypsin was the enzyme used in the digestion. Parent mass error tolerance was set to "2 Da" because this was low resolution, low mass accuracy ion trap data. The proteome.dta file was uploaded and the file format was set to "dta", because these were concatenated "dta" files. The dta file is a file that represents a single MS2 or fragment mass spectrum. The dta is basically a list of mass vs. intensity data pairs, it is all that is needed to recreate the MS/MS mass spectrum. Here is a single dta file that you can look at or download to see what the basic file structure is like. Click on the dta link to see the file format. In a dta file the first line contains the parent mass and the assumed charged state. This first line is followed by the list of mass and intensity pairs. These files are used to search a sequence database with MS data. If you have a lot of these files, as with a proteomics dataset, they can be concatenated for upload, or Phenyx will accept zip or tar.gz dta files. We have already concatenated these files for you in the file, proteome.dta.
Figure 3.
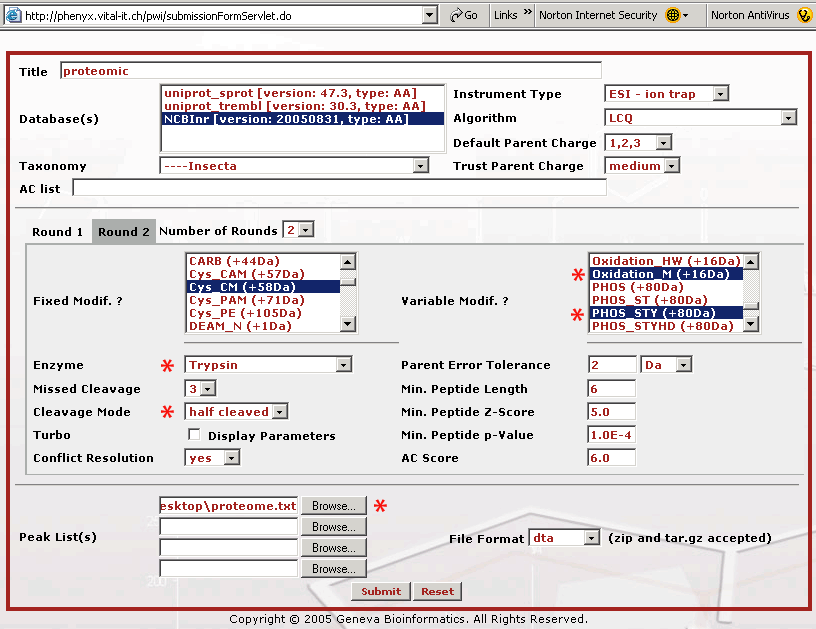
Figure 3. This screenshot shows the parameter entry page for the Round 2 search. If you are following along, you will need to press the Round 2 button to get to this page. The Round 2 search page allows you to pick multiple modifications for the second round search. Bear in mind that the modification(s) selected in the first Round also need to appear in the second Round, as fixed or variable modification(s), depending on your confidence level. With most sequence search engines if you pick many variables the search can take days to complete. The way the second round search works is that the main protein players are identified in the Round 1 search, where few modification variables were defined. The Round 1 search can be done relatively quickly. Once these proteins are identified, Phenyx makes a subset database that can be searched very quickly with a broader set of modification variables. Once the parameters for the Round 1 and 2 searches are selected, the entire search operation becomes transparent and very automatic. Once you have selected your Round 2 search parameters, press the "Submit" button to start the search. The choices we made above will allow for an unprecedented 3 missed cleavages, plus it will pick up peptides that have only one trypsin end, K/R, half tryptic. We have also chosen to look for every possible serine, threonine, and tyrosine phosphorylation. We have found that Phenyx will save your variable selections which can be handy. However, be careful when performing new searches that your choices are correct, for example a modification variable can hide out of view on the list. To be absolutely certain that your settings are correct hit the "Reset" button at the bottom of the page and re-pick your mods when making a new search.
Figure 4.
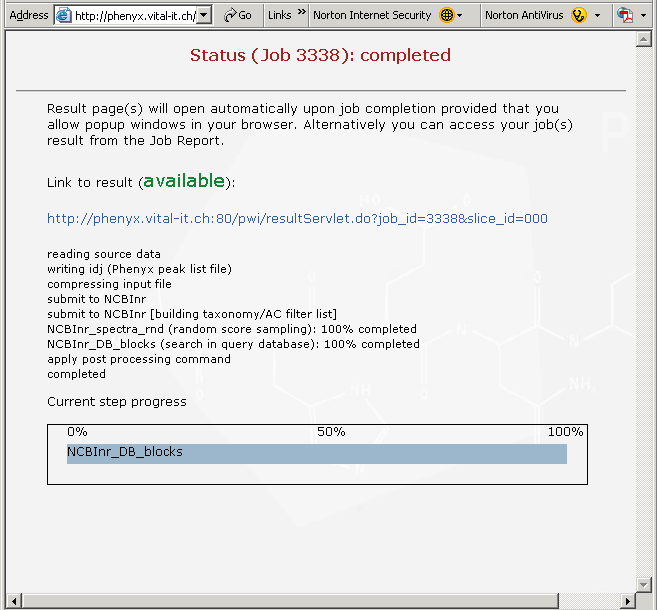
Figure 4. This is the page that appears when your search is complete. Since this is a rather large proteomics file, the search will take a few minutes. Click on the link in this view, if you are following along, to see your results. If you have pop-ups blocked in your browser it may interfere with this page, we have found that you can recover by just hitting the refresh button on your browser.
Figure 5.
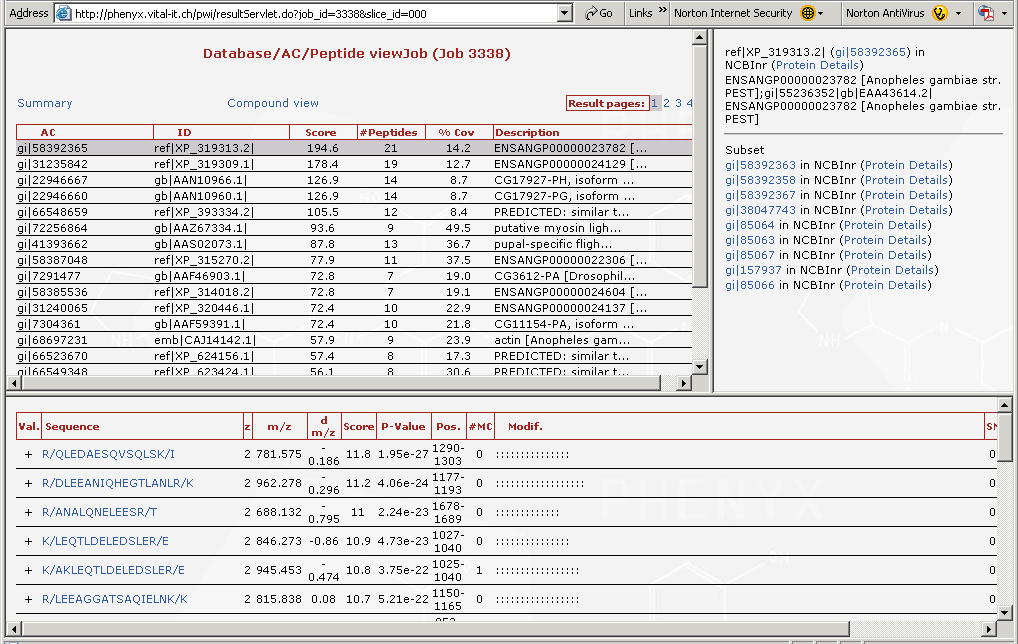
Figure 5. Here are the proteins that were identified. The list of "gi" numbers in the top left hand frame are all of the proteins that were found. Note the scroll bar, there are many more proteins on this list. These were the proteins from a mosquito that flew into the lab one day, and lit on a technicians arm. The mosquito was tipped into an micro tube and smashed in the presence of 8M urea, then reduced and alkylated. When you highlight a protein in the "gi" list, in the top frame you can access the protein details by following the link in the upper right frame labeled NCBInr (Protein Details). The protein details page is split and is shown in Figures 6 and 7.
Figure 6.
 \
\
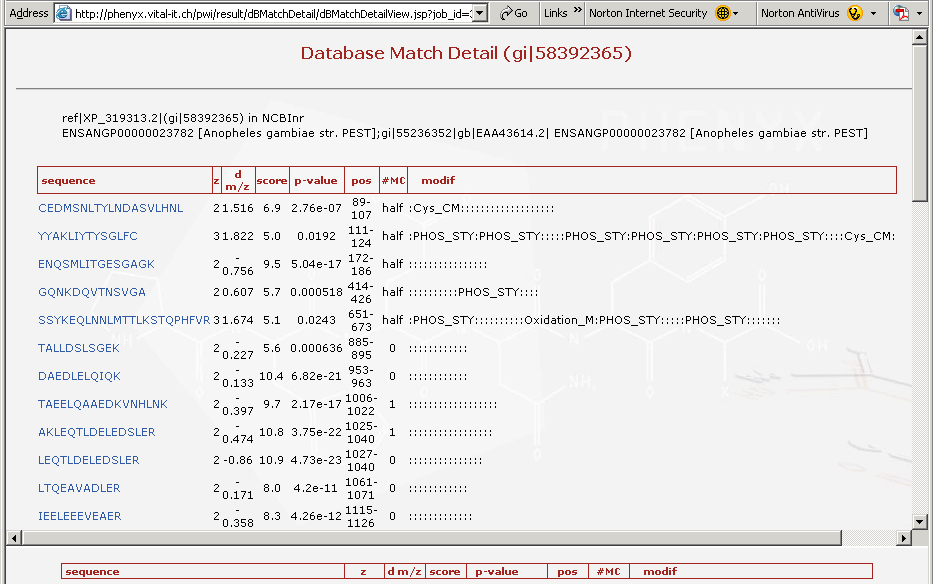
Figure 6. This is the top portion of the protein details page. The top frame shows all of the peptides identified for that particular protein.
Figure 7.
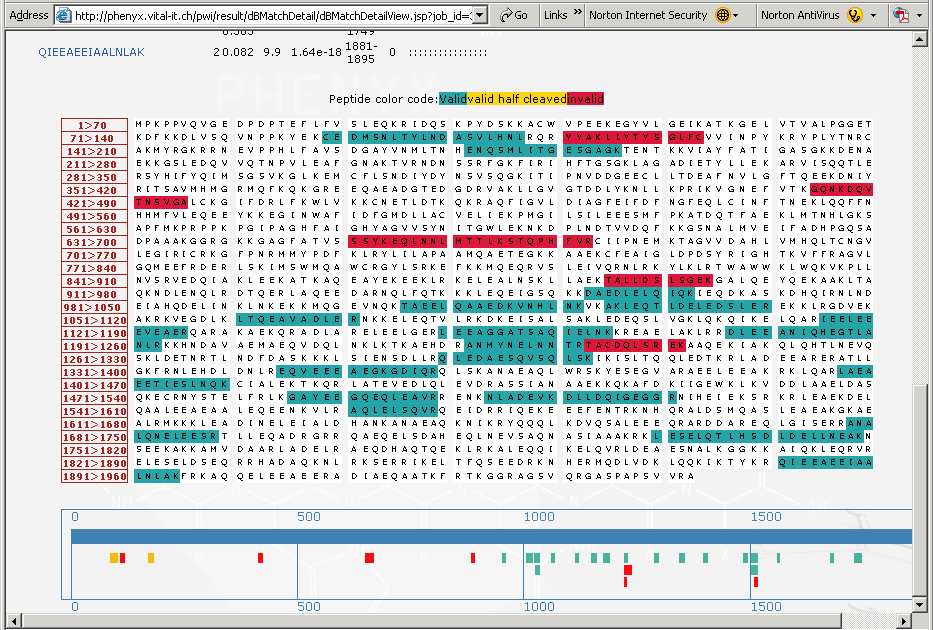
Figure 7. This is the lower portion of the “protein details” page that shows the coverage map of the protein identified. The peptides, highlighted in the "blue green" color are the peptides that have been validated by Phenyx. The peptides highlighted in red are those peptides that were correlated but deemed to be invalid. Below the sequence coverage map in Figure 7, above, is a cartoon of the coverage map showing in detail all of the peptides identified, notice that some half cleaved peptides (in yellow) were identified and validated.
Figure 8.
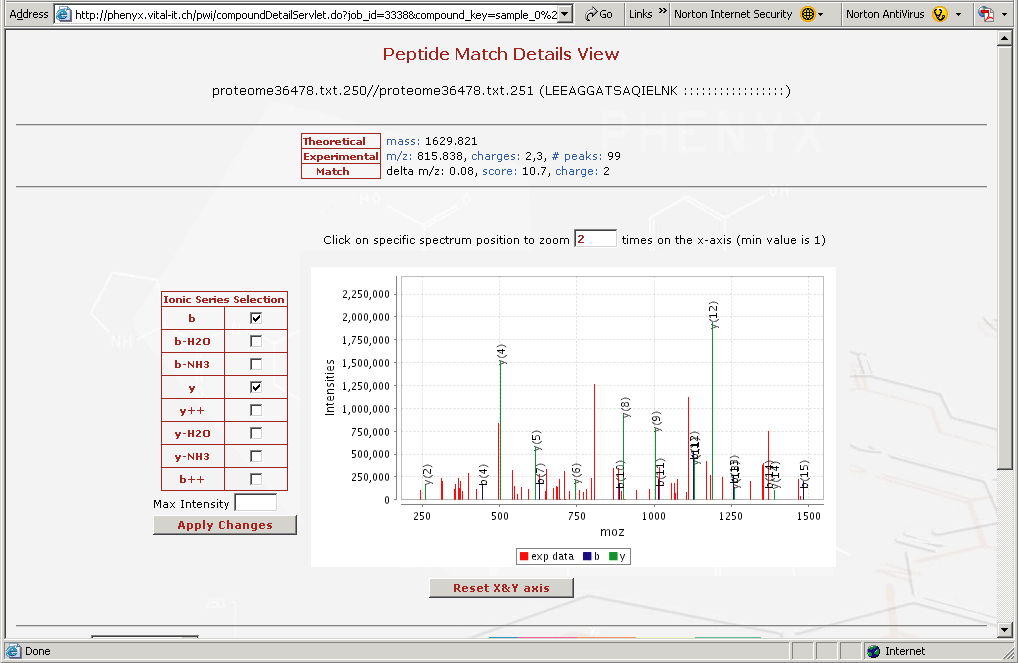
Figure 8. This is the "Peptide Match Details View" This view can be accessed by clicking on any of the identified peptides in the results view in Figure 6.
Figure 9.
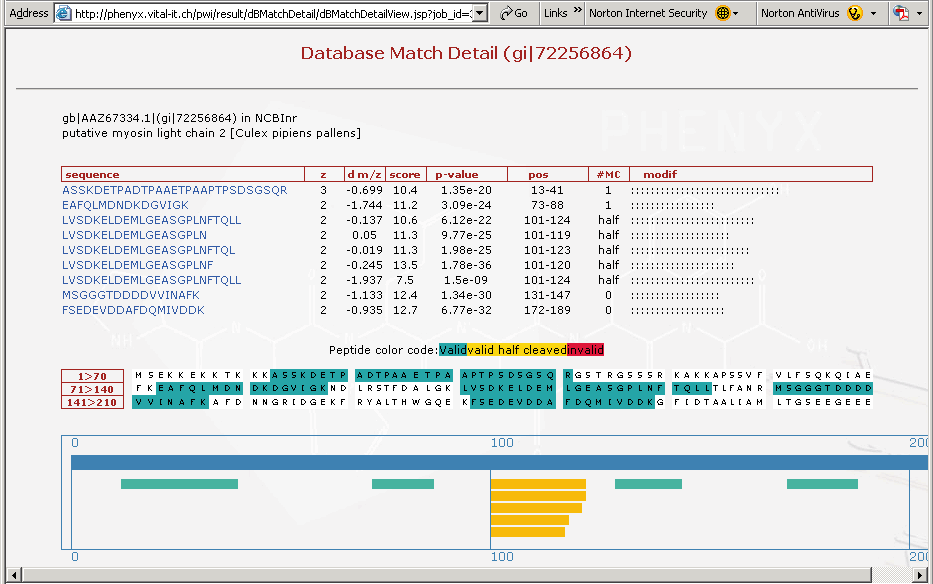
Figure 9. This is a screenshot showing the third protein on the list, just to demonstrate how important the "half cleaved" option in the Round 2 search can be. In Figure 9 above you can see the cluster of half cleaved peptides identified in the search for this particular protein, see yellow peptides in bottom frame of Figure 9. The amino terminal ends for these peptides are all the same, and are tryptic. The carboxyl terminus of these yellow peptides appear to be differentially processed. These peptides may have been processed by an insect enzyme or may have occurred in the sample work-up, although if you remember the sample preparation we described involved no storage or freeze thaw of the sample. It is likely that this cleavage pattern is an insect related phenomenon and happened In-Vivo. This could be a very interesting result and demonstrates that one needs to search with more than just strict trypsin specificity.
Figure 10.
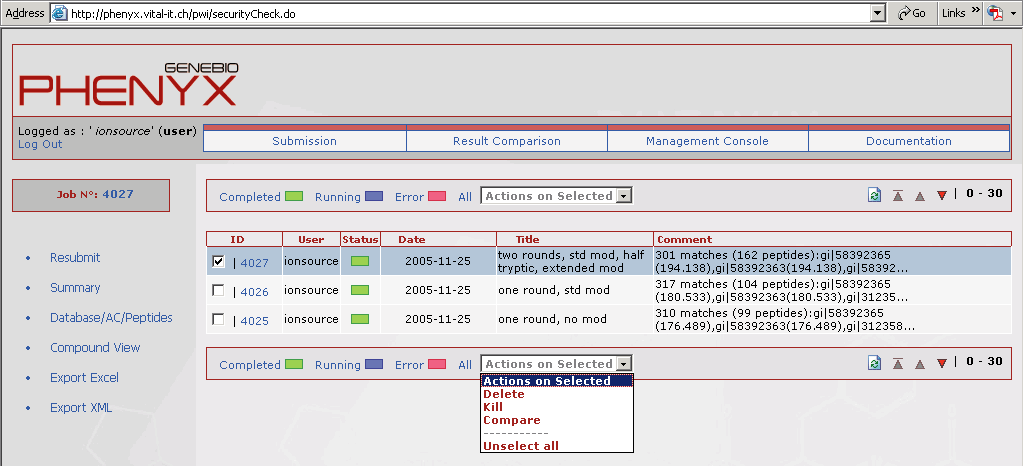
Figure 10. This view shows the results comparison page that can be accessed through the account home page. It is very easy to compare search jobs. Just type the job number in the "add job" box. To compare multiple jobs just separate the job numbers with a comma. Even Sequest and Mascot results can be displayed in this view!
Figure 11.

Figure 11. This screen shot is a return to the Phenyx home page. You can see all of the searches we have performed. These searches can be kept, deleted, reviewed and compared.
Figure 12.
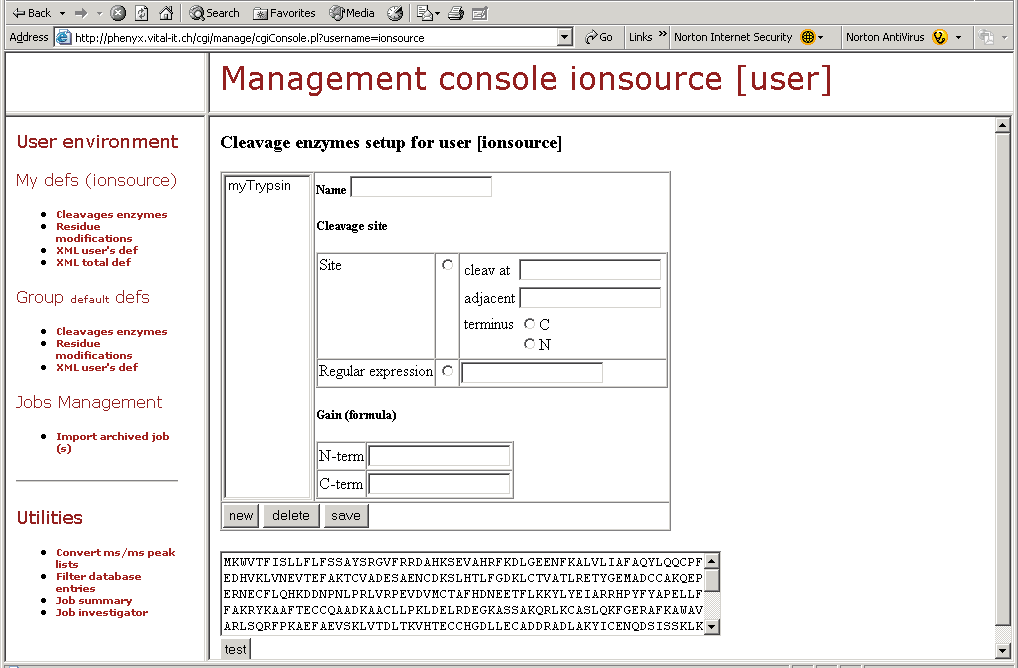
Figure 12. This screenshot shows one part of the "Management Console." Here you can design your own mods, define or modify cleavage specificities, and perform many other amazing tasks as shown by the topics in the left hand frame of this figure.
Conclusion:
(Disclaimer: These facts were true to the best of our knowledge at the time of review, 11/20/2005. These points may change as the field of proteomic searching evolves. Feel free to e-mail webmaster@ ionsource.com with all corrections.)
The public version of Phenyx provides many special features not found in other on-line sequence search engines. We have listed the features below that we think make Phenyx special. Many of these features are unique to Phenyx and are unprecedented in the realm of free on-line search engine resources.
Here is a list of features that we found valuable. We have starred in red the features that we found or believe to be unique among on-line sequence search engines.
1.) A big plus for Phenyx is no sample submission limit.
2.) * The ability to search a selected subset or even a single protein in a targeted search, using the AC list function is unique.
3.) * The ability to create an account and save past submissions for later review is unique.
4.) * The ability to compare searches side by side, to do comparative proteomics, is unprecedented from a free on-line resource.
5.) * The ability to import Mascot and Sequest results to compare against each other or the Phenyx result is also unprecedented in an on-line resource.
6.) The automated second round search function is also shared by X!tandem.
7.) * Not apparent is the reverse database search that is performed transparently with every Phenyx search to aid in the probability calculation. We think that this is unique among on-line search engines.
8.) * Spectral correlation conflict resolution is unique to Phenyx.
9.) * The on-line
"Management Console" function of Phenyx is truly unique, providing
many useful tool for protein identification. The only thing missing in the
Pheny public version is the ability
to upload personal sequence databases, which no one has, in a free on-line
search engine. (but is available in the Phenyx local version)
10.) * The ability to upload 4 search jobs simultaneously is a unique feature to an on-line resource.
11.) Another important feature is that results can be exported as an Excel or XML file.
12.) * Large groups of dta files can be uploaded in "zipped" folders or .tar files, without concatenation, this is unique. (this feature was just brought to our attention, and we have not tested it.)
13.) When searching with Phenyx one also has the ability to choose multiple databases. This can be very handy, for example, if you are searching a plant database and also want to search a fungal database.
e-mail webmaster@ionsource.com
with all inquiries
home | terms
of use (disclaimer)
Copyright © 2005-2016, IonSource, LLC, All rights reserved.
Last updated: Tuesday, January 19, 2016 02:51:47 PM