| How to download and use ReAdW | ||
| IonSource.com | ||
|
Implementing the ISB program ReAdW for the conversion of Xcalibur raw files to the new universal mzXML format. Your new mzXML file can then be viewed with the free Insilicos viewer and searched on the public version of the Phenyx . |
||
|
(Even though this software download did not cause us any
trouble our policy is to never download software from the
internet onto a "mission critical
computer." Also, the software described here is independent of
IonSource so please read our terms of use
disclaimer and theirs)
|
||
|
Download the latest version of "ReAdW"
|
||
|
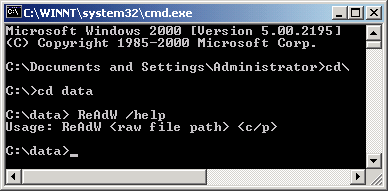
Place the ReAdW program in the same
folder as the Xcalibur raw data file, see Figure 1, bottom. You will need to run
ReAdW from the "RUN" dialog see top gif of Figure 1. ReAdW is
easy to download and to run. Download ReAdW to the folder that
contains your data file. 1.) Go to the START menu on your
computer, and click on the "Run..." tab. |
||
|
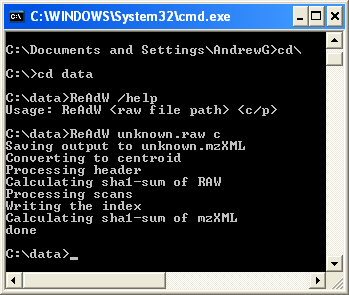
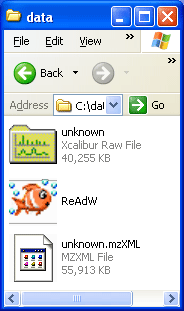
Here is a screen shot of the run dialog and the folder. Notice in the folder view that the created mzXML file is 1.4 times bigger. |
||
|
Figure 1
To run the program type "ReAdW datafile name c" and hit return and wait until you see "done" as shown above.
|
||
|
Download the free Insilicos Viewer at http://www.insilicos.com/products.html |
||
|
Figure 2 |
||
|
The free viewer comes with an installer and installs painlessly. It is a small program and very efficient so it launches quickly and also opens files quickly. |
||
|
Once the viewer is installed open the program and open the file you just converted to mzXML format. It is as easy as that! It is a revolution I think. Now anyone can read native Finnigan Xcalibur files. Thank you Insilicos and ISB! Here are a few screen shots of the program. |
||
|
Figure 3 |
||
|
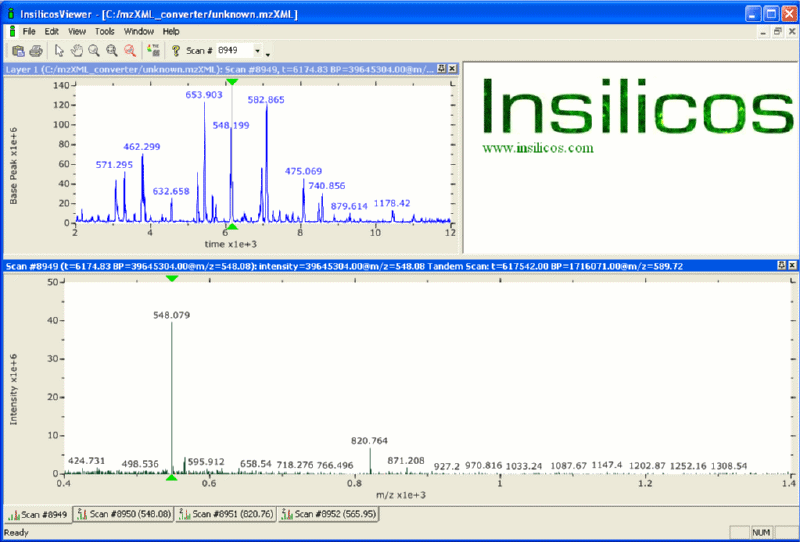
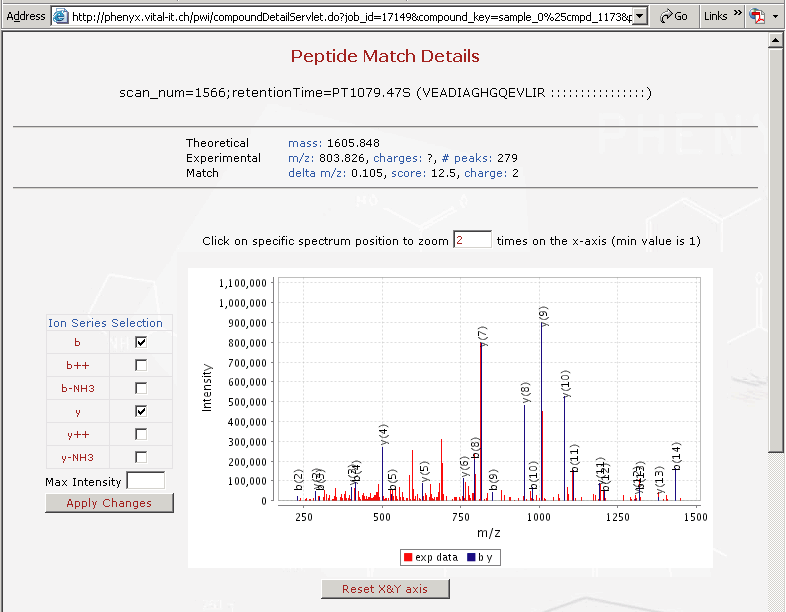
Figure 3: This is a screen shot of the viewer with a base peak plot in the upper left hand frame and a full MS spectrum in the bottom plot. Notice the tabs at the bottom, the first tab is the full MS followed by the three MS2s that were performed. |
||
|
Figure 4 |
||
|
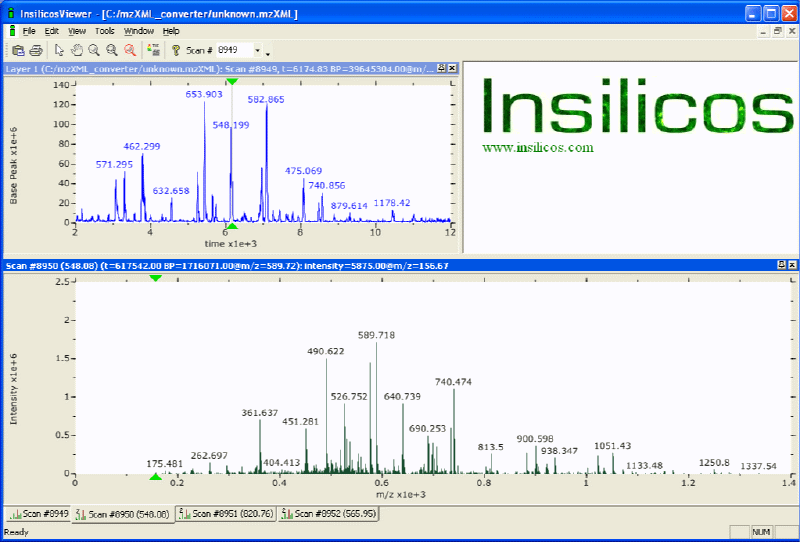
Figure 4: Shows a screen shot of the viewer with a base peak plot in the upper left hand frame and an MS2 spectrum in the bottom plot. |
||
|
Now that you have your data in the new common file format, mzXML, you can submit your files to those proteoimc search engines that accept mzXML. If your vendor does not support mzXML tell them that you want it. |
||
| One of the
search engines that accepts mzXML is Phenyx from GeneBio. You can
download the Public Phenyx and submit your new mzXML file
for protein database searching. Follow this link
to the Phenyx public search engine.
|
||
|
Figure 5 |
||
|
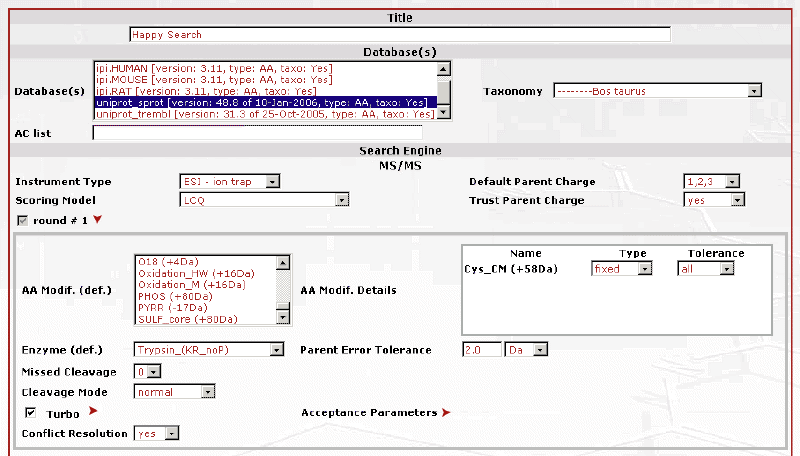
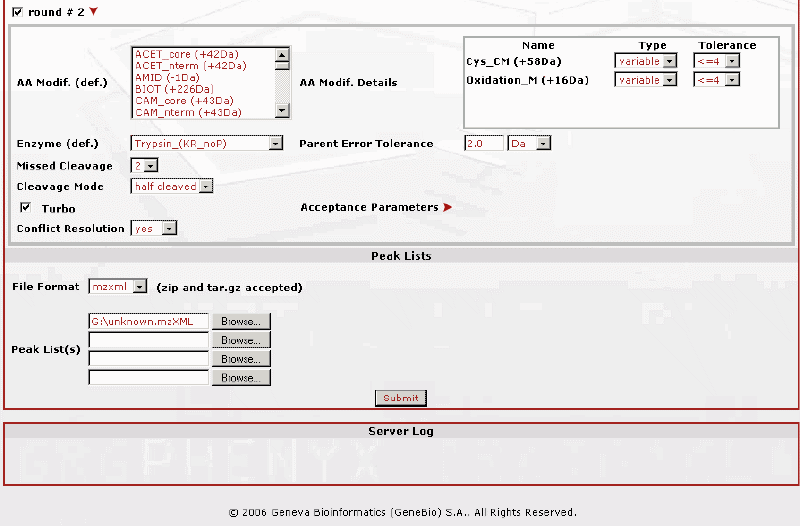
Below is the second part of the submission page. Notice that there is an option for a second round search in which you can include more variables. |
||
 |
||
|
|
||
|
Figure 6
|
||
|
|
||
|
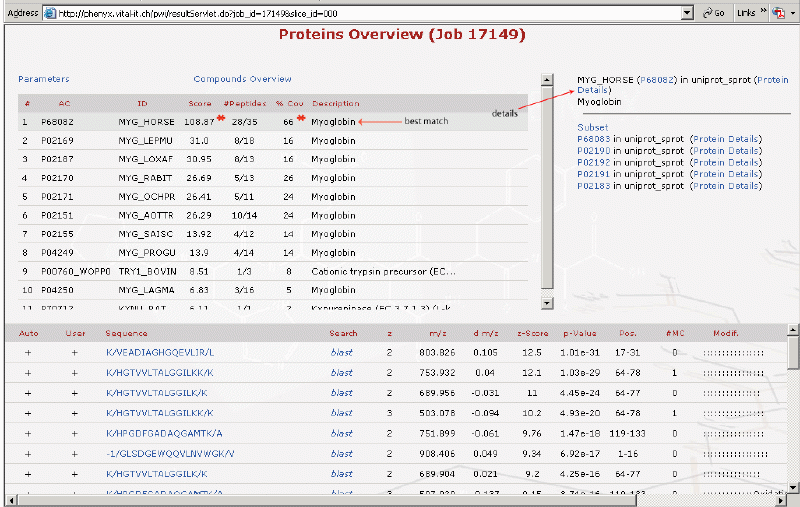
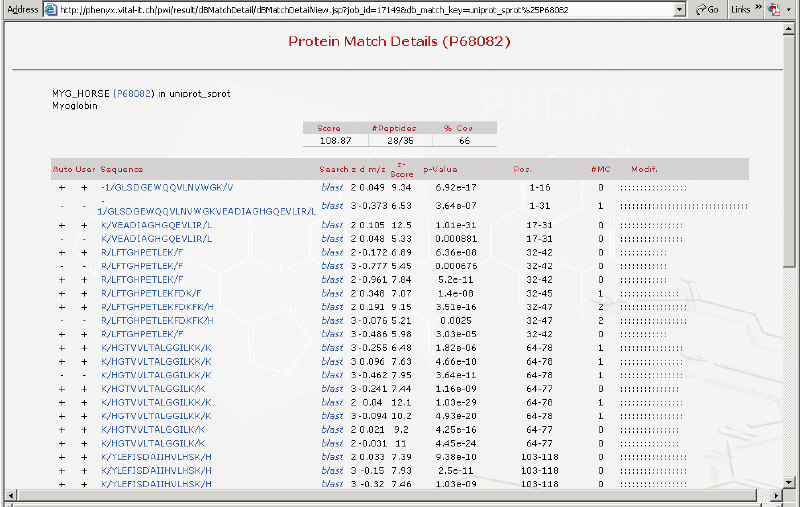
This is the protein match details page. All of the peptides identified are listed here as well as the protein sequence coverage map.
|
||
|
|
||
|
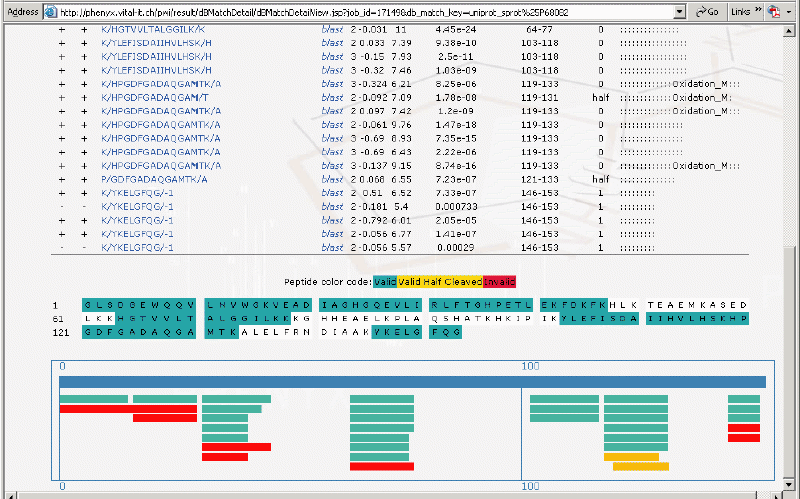
Shown below is the sequence coverage map. Notice the half cleaved peptides and those matches that were deemed invalid.
|
||
|
Figure 7
|
||
|
|
||
| Conclusion: A common file format has been a dream of mass spectrometrists since the second instrument company made a mass spectrometer. mzXML has advanced that dream, it is our responsibility to encourage vendors to support this format. Thanks to ReAdW.exe we can now give our clients native data files and point them to ISB or Insilicos to get the converter and the viewer. I have every confidence that this type of open source effort will speed up scientific research. "Enable the researcher" should be our motto. Some search engine sellers or MS vendors may fear this advance, but what it will really mean is that more people will have access to MS data and more people will be able to search their own data, and hence more search engine software will be sold. It will be good for everyone. | ||
| e-mail the webmaster@ionsource.com with all inquiries | ||
| home
| terms of use
(disclaimer) Copyright © 2004-2016 IonSource, LLC All rights reserved. Last updated: Tuesday, January 19, 2016 02:51:41 PM |
||