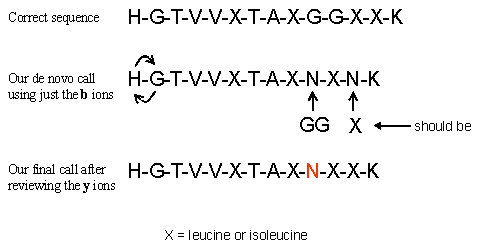|
Notice: Advertisement |
||
|
-
|
||
|
-De Novo Peptide Sequencing Tutorial 1st De Novo Exercise the answer page
|
||
|
The correct sequence for this peptide is,
HGTVVLTALGGILK or more accurately the best that low energy CID can do HGTVVXTAXGGXXK
The first thing that we did was to do the math to determine the penultimate b ion. If we could determine the C-terminal residue from the observed immonium ions we could calculate the penultimate b ion using the formulas below. 1379.4 - 18 -156 = 1205.4 for Arginine 1379.4 - 18 -128 = 1233.4 for Lysine Since we could not observe the low end of the ion trap spectrum we had to do the math for both likely tryptic peptide C-terminal residues. We found that the C-terminal residue matched for Lysine. The b ion series extended all of the way into the low end of the spectrum. The a ion type fragments did accompany some of the b ions down the spectrum giving us some confidence that we were following the correct sequence. For the b ion series, once we got down to the low end of the spectrum we had to use mass and amino acid residue combinations to determine the last two amino terminal residues. From the observed b ion series we could not determine the order of the remaining two amino terminal amino acids. Once the b ions were determined we calculated the expected y ion series using the formula [M+H]+ - observed b ion + 1 = corresponding y ion. We then went through and redid the delta masses on all of the calculated and observed y ions, obtaining the y ion sequence. The y ions were more abundant and let us determine the order of the two amino terminal amino acids as well as removing the N-X ambiguity that we observed in the b ion series. We had this N-X ambiguity because the b ion series was much less abundant that the y ion series, and a more accurate mass was easier to determine for the y ion series With the b ion series, we still missed the G-G sequence and substituted an asparagine, a rookie mistake. Here is the sequence that we determined (called) from both the b and y ion series.
All considered, we feel that we did pretty good. We did mistake the double glycine for an asparagine. The peak that would have indicated the glycine in the b ion series was very small and we fell into the N = GG trap. Once this mistake was made we missed the glycine peak in the y ion series even though the peak was fairly observable. Even though one can calculate the corresponding y or b ion once a b or y ion is observed we would suggest calling both the y and b series independently to remove any bias. This would have saved us from calling GG as an N. Perhaps we could do better with q-TOF data, let's see as we proceed onto the next exercise. Still, we did pretty good; hope you did too. Don't let anyone tell you that trap data is bad for de novo, you just need to try a little harder and be a little wary. If you would like to see the scribbling of a mad man, as I marked my spectrum up in this exercise, you can down load this pdf.
|
||
|
|
||

|

|
|
|
|
||
|
|
||
|
|
||
| e-mail the webmaster@ionsource.com with all inquiries | ||
| home
| terms of use
(disclaimer) Copyright � 2012 IonSource All rights reserved. Last updated: Monday, February 01, 2016 11:05:38 AM |
||
|
|
||