-
Peptide-Mass Fingerprinting

MALDI TOF Mass Spectrum
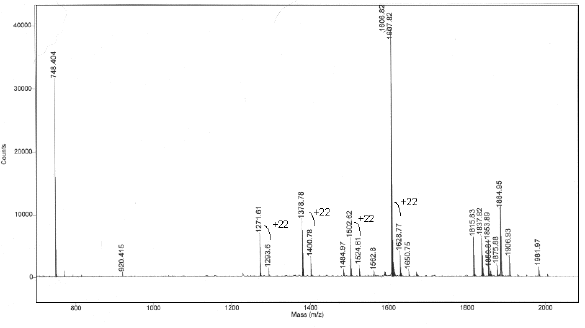
Discussion:
See references 10-11 for more information on the Aldente search engine. Be sure to cite Aldente if you use any future results in a publication. How to cite
Now that you have mastered peptide-mass fingerprinting try your hand at a technique called "Sequence Tag" on the following page.
-
-
Henzel, W. J.; Stults, J. T.; Watanabe, C. Proceedings of the Third Symposium of the Protein Society; Seattle, WA, 1989.
-
Henzel WJ, Billeci TM, Stults JT, Wong SC, Grimley C, Watanabe C. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5011-5.
-
Mann M, Hojrup P, Roepstorff P. Use of mass spectrometric molecular weight information to identify proteins in sequence databases. Biol Mass Spectrom. 1993 Jun;22(6):338-45.
-
Pappin DJ, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting.
Curr Biol. 1993 Jun 1;3(6):327-32. (first to coin the term) -
James P, Quadroni M, Carafoli E, Gonnet G. Protein identification by mass profile fingerprinting.
Biochem Biophys Res Commun. 1993 Aug 31;195(1):58-64. -
Yates JR 3rd, Speicher S, Griffin PR, Hunkapiller T. Peptide mass maps: a highly informative approach to protein identification. Anal Biochem. 1993 Nov 1;214(2):397-408.
-
Jensen ON, Podtelejnikov AV, Mann M. Identification of the components of simple protein mixtures by high-accuracy peptide mass mapping and database searching. Anal Chem. 1997 Dec 1;69(23):4741-50.
-
Clauser KR, Baker P, Burlingame AL.Role of accurate mass measurement (+/- 10 ppm) in protein identification strategies employing MS or MS/MS and database searching.
Anal Chem. 1999 Jul 15;71(14):2871-82. PMID: 10424174 -
Henzel WJ, Watanabe C, Stults JT. Protein identification: the origins of peptide mass fingerprinting. J Am Soc Mass Spectrom. 2003 Sep;14(9):931-42.
- Tuloup M, Hernandez C, Coro I, Hoogland C, Binz P-A, Appel R D, Aldente and BioGraph: An improved peptide mass fingerprinting protein identification environment, Swiss Proteomics Society 2003 Congress: Understanding Biological Systems through Proteomics, Basel, Switzerland, 2-4 Dec. 2003, Ed. FontisMedia (ISBN 2-88476-004-0), 174-176
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins M R, Appel R D, Bairoch A, Protein Identification and Analysis Tools on the ExPASy Server, in: The Proteomics Protocols Handbook (in press, 2005 Feb.). Edited by John M. Walker, Humana Press, New Jersey.

Copyright � 2004-2016 IonSource All rights reserved.
Last updated: Tuesday, January 19, 2016 02:48:34 PM
